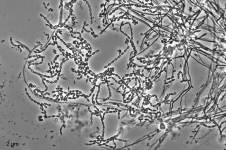
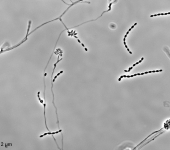
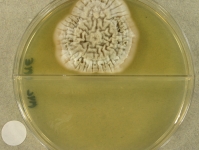
Search Details
| UAMH Number: | 9571 |
|---|---|
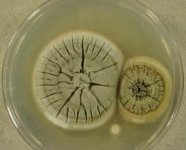
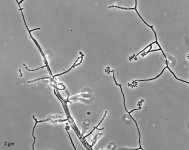
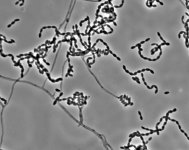
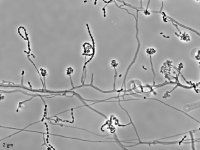
| Species Name: | Sagenomella sp. |
| Type: | |
| Synonyms: | |
| Taxonomy: | FUNGI Ascomycota, Eurotiomycetes, Eurotiales, Trichocomaceae |
| Strain History: | McCracken, B. (2) -> UAMH |
| Substrate: | roots of Vaccinium membranaceum | Location: | USA Idaho, Priest Lake (GEO: 48.588,-116.867) |
| Isolator: | B. McCracken (PL1DdiE) |
| Isolation Date: | |
| Date Received: | 1999-06-23 |
| Characters: | CYCLOHEXIMIDE sensitive (MYC) // MESOPHILIC NG @ 35C // MOLECULAR SYSTEMATICS ITS phylogenetic analysis groups with Talaromyces ocotl (Sagenomella anamorph) // MOLECULAR SYSTEMATICS relationship to Phialosimplex within the Trichocomaceae - Sigler L, Sutton DA, Gibas CF, Summerbell RC, Noel RK, Iwen PC, Med Mycol 48:335-345, 2010 // ODOR/ ODOUR sour |
| Compounds: | |
| Cross Reference: | CBS 113280 |
| Pathogenic Potential: | Human: unknown | Animal: no | Plant: no |
| Biosafety Risk Group: | RG1 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide a CFIA Written Authorization for transfer (http://www.inspection.gc.ca/plants/plant-pests-invasive-species/directives/date/d-12-03/eng/1432656209220/1432751554580#app2) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. This strain is not available for shipment to Cuba, the Democratic People's Republic of Korea, Iran or Syria. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 9773 |
| Sequences: | >UAMH09571_D1D2 TGCCCTAGTAACGGCGAGTGAAGCGGCAACAGCTCGAATTTGAAATCTGGCTTCGGCCCGAGTTGTAATCTGTAGAGGACGCTTTTGGCGCGGTGCCTTCCGAGTTCCCTGGAACGGGACGCCATAGAGGGTGAGAGCCCCGTAAGGTCGGACACCAAGCCTCTGTAAAGCTCCTTCGACGAGTCGAGTAGCTTGGGAATGCTGCTCTAAGCGGGAGGTAAACTTCTTCTAAAGCTAAATACTGGCCAGAGACCGATAGCGCACAAGTAGAGTGATCGAAAGATGAAAAGCACTTTGAAAAGAGGGTCAAATAGTACGTGAAATTGTTGAAAGGGAAGCGCTCATGACCAGACTTGCGCCTGGCGGATCATCCGGCTGTCTCAGCCGGTGCACTCCGCCTGGCGCAGGCCAGCGTCGGCTGTTGCGGGGGGACAAAAGCACTGGGAACGTAGCTCTCCTCGGAGAGTGTTATAGCCCTATGCAGAATACCCCCGCGGCGGCCGAGGTCCGCGCTCTGCAAGGACGCTGGCGTAATGGTCATCAGCGACCCGTCTTGAAACACGGACCAAGGAGTCGAGGTTTTGCGCGAGTGTTCGGGTGCAAAGCCCCAGCGCGTAATGAAAGTGAACGCAGGTGAGAGCTTCGGCGCATCATCGACCGATCCTGATGTATTCGGATGGATTTGAGTAGGAGC >UAMH09571_LSU TGCCCTAGTAACGGCGAGTGAAGCGGCAACAGCTCGAATTTGAAATCTGGCTTCGGCCCGAGTTGTAATCTGTAGAGGACGCTTTTGGCGCGGTGCCTTCCGAGTTCCCTGGAACGGGACGCCATAGAGGGTGAGAGCCCCGTAAGGTCGGACACCAAGCCTCTGTAAAGCTCCTTCGACGAGTCGAGTAGCTTGGGAATGCTGCTCTAAGCGGGAGGTAAACTTCTTCTAAAGCTAAATACTGGCCAGAGACCGATAGCGCACAAGTAGAGTGATCGAAAGATGAAAAGCACTTTGAAAAGAGGGTCAAATAGTACGTGAAATTGTTGAAAGGGAAGCGCTCATGACCAGACTTGCGCCTGGCGGATCATCCGGCTGTCTCAGCCGGTGCACTCCGCCTGGCGCAGGCCAGCGTCGGCTGTTGCGGGGGGACAAAAGCACTGGGAACGTAGCTCTCCTCGGAGAGTGTTATAGCCCTATGCAGAATACCCCCGCGGCGGCCGAGGTCCGCGCTCTGCAAGGACGCTGGCGTAATGGTCATCAGCGACCCGTCTTGAAACACGGACCAAGGAGTCGAGGTTTTGCGCGAGTGTTCGGGTGCAAAGCCCCAGCGCGTAATGAAAGTGAACGCAGGTGAGAGCTTCGGCGCATCATCGACCGATCCTGATGTATTCGGATGGATTTGAGTAGGAGCGTAAAGCCTCGGACCCGAAAGATGGTGAACTATGCCTGTATAGGGTGAAGCCAGAGGAAACTCTGGTGGAGGCTCGCAGCGGTTCTGACGTGCAAATCGATCGTCAAATATGGGCATGGGGGCGAAAGACTAATCGAACCATCTAGTAGCTGGTTACCGCCGAAGTTTCCCTCAGGATAGCAGTGTTGATCTTCAGTTTTATGAGGTAAAGCGAATGATTAGGGACTCGGGGGCGCTTATTAGCCTTCATCCATTCTCAAACTTTAAATATGTAAGAAGCCCTTGTTACTTAGCTGAACGTGGGCCTTCGAATGTAACAACACTAGTGGGCCATTTTTGGTAAGCAGAACTGGCGATGCGGGATGAACCGAACGCGGGGTTAAGGTGCCGGAGTGGACGCTCATCAGACACCACAAAAGGCGTTAGCACATCTTGACAGTAGGACGGTGGCCATGGAAGTCGGAATCCGCTAAGGACTGTGTAACAACTCACCTACCGAATGTGCTAGCCCTGAAAATGGATGGCGCTCAAGCGTCCCACCCATACCCCGCCCTCAGGGTAGTAGCGATGCCCTGAGGAGTAGGCGGCCGTGGGGGTCAGTGACGAAGCCTAGGGCGTGAGCCCGGGTCGAACGGCCCCT >UAMH09571_GQ169325_SSU-LSU TCATTACCGAGTGAGGGTCCCTTGCGGGCCCAACCTCCCACCCGTGTTTAACTATACCGTGTTGCTTCGGCGGGCCCACTGGGGCCTGTTCCCGGTCGCCTGGGGGGGTGAAACCCCCCGGGTCCGTGCCCGCCGGAGACCCCTTGAACCCTGAGTGAATCGAGTGTCGTCTGAGTTTGAATTAAATCATTAAAACTTTCAACAACGGATCTCTTGGTTCCGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCCGTGAATCATCGAATCTTTGAACGCACATTGCGCCCCCTGGCATTCCGGGGGGCATGCCTGTCCGAGCGTCATTGCTACCCTCAAGCGCGGCTTGTGTGTTGGGCGCTGTCCCCCCGGGGACAGGCCTGAAAGGCAGTGGCGGCGTCGCGTCCGGTCCTCGAGCGTATGGGGCTTTGTCACTCGCTCTGTGGGGTCCGGCCGGGGCCTGTCTACCCAATCTTTTCTCAAGGTTGACCTCGGATCAGGTAGGGATACCCGCTGAACTTAAGCATATCAATAAGCGGAGGAAA |
IMAGES: