<< back to search
IMAGES:
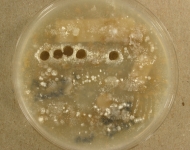
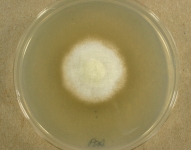
Search Details
add to cart
| UAMH Number: | 7712 |
|---|---|
| Species Name: | Nannizziopsis mirabilis |
| Type: | Nannizziopsis mirabilis |
| Synonyms: | |
| Taxonomy: | FUNGI Ascomycota, Eurotiomycetes, Onygenales, Nannizziopsidaceae |
| Strain History: | Udagawa, S.-I. (SIU 5954) -> UAMH |
| Substrate: | soil sample | Location: | USA New Jersey (GEO: 40.058,-74.406) |
| Isolator: | |
| Isolation Date: | 1993 |
| Date Received: | 1994-11-15 |
| Characters: | CULTURE CONDITIONS ascomata produced on OAT after 2 mos - fide UAMH 2011 // MOLECULAR SYSTEMATICS excluded from Nannizziopsis clade - Sigler L et al 2013 // MOLECULAR SYSTEMATICS phylogeny of Nannizziopsis and related onygenalean fungi - Sigler L, Hambleton S, Pare JA, J Clin Microbiol 51:3338-3357, 2013 // SYSTEMATICS/ TAXONOMY description - Uchuyama S,Kamiya S, Udagawa S-I, Mycoscience 36:205-209, 1995 (Click for publications citing UAMH 7712) |
| Compounds: | |
| Cross Reference: | |
| Collections: | Living Strains; Dried Herbarium Material |
| Pathogenic Potential: | Human: no | Animal: yes | Plant: no |
| Biosafety Risk Group: | RG2 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) and a CFIA Written Authorization for transfer (http://www.inspection.gc.ca/plants/plant-pests-invasive-species/directives/date/d-12-03/eng/1432656209220/1432751554580#app2) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. This strain is not available for shipment to Cuba, the Democratic People's Republic of Korea, Iran or Syria. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 413562 |
| Sequences: | >UAMH07712_KF477243_ITS GCGCCCGCCAGTGGAACTTTACCTGAACTTTTGAAAAGTGGCCGTCTGAGTGATTTTTACGAATCAATCAAAACTTTCAACAACGGATCTCTTGGTTCCGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAGTGTGAATTGCAGAATTCCGTGAATCATCGAATCTTTGAACGCACATTGCGCCCTCTGGTATTCCGGGGGGCATGCCTGTCCGAGCGTCATTGCAACCCTCAAGCGCGGCTTGTGTGATGGGTCCCGTTGTCTCCCGCTCCCGGCGAGACAGGCCCGAAATGCAGTGGCGGCGTCCCGAATCGGGTGCCTGAGTGAATGGGATTTATACATCCGCTCGAGCGGCCCCGTCGGCGCCGGCCGGTCATCCAATTACCAATCGG |
IMAGES: