<< back to search
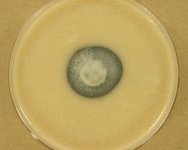
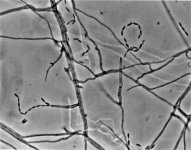
IMAGES:
Search Details
add to cart
| Strain Number: | UAMH 4935 |
|---|---|
| Species Name: | Phialophora oxyspora |
| Type: | Phialophora oxyspora |
| Synonyms: | Cyphellophora oxyspora |
| Taxonomy: | FUNGI Ascomycota, Eurotiomycetes, Chaetothyriales, Cyphellophoraceae |
| Strain History: | CBS 698.73 -> UAMH |
| Substrate: | aphids in Clerodendron monahassa | Location: | SRI LANKA (GEO: 7.873,80.772) |
| Isolator: | |
| Isolation Date: | |
| Date Received: | 1984-03-05 |
| Characters: | MOLECULAR SYSTEMATICS relationships among Cyphellophora and related Phialophora species - Feng P, Lu Q, Najafzadeh MJ, van den Ende AHG et al, Fungal Diversity epub 15-Aug-2012 |
| Compounds: | |
| Cross Reference: | CBS 698.73 |
| Pathogenic Potential: | Human: yes | Animal: yes | Plant: no |
| Biosafety Risk Group: | RG2 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 319853 |
| Sequences: | >UAMH04935_JQ766375_beta-tubulin GTGAGCCGTCATTCTCGTCGTCGCCCCTCTTCACCGCCTAATCAGTTGCAGGCAACTCATCTCCGGCGAGCACGGTCTCGATGGCTCCGGCAAGTAAGTCAATATCCCGAATCTCGTCGAGTTCGAGCTTTGCTGACCTTAACCTAGCTACAATGGCACTTCGGACCTCCAGCTCGAGCGTATGAACGTGTACTTCAATGAGGCAAGTCGCAATGACCCACTAGGCAGCCTGGCGCTAACATCGACGTTAGGCCGCTGGCAACAAGTATGTTCCTCGCGCCGTCCTCGTCGATCTCGAGCCCGGTACCATGGATGCCGTCCGTGCCGGCCCTTTTGGACAGCTGTTCCGTCCCGACAACTTCGTGTTTGGTCAGAGCGGCGCCGGTAA >UAMH04935_JQ766498_LSU CGGCAACAGCTCAAATTTGAAATCTGGCCTCTTTGGGGTCCGAGTTGTAATTTGGAGAGGATGTTTCGGGCACGGTCCGGGCTTAAATTTCTTGGAACAGAATGTCACAGAGGGTGAGAATCCCGTCTGGAGTCCGCGACACGACCTATGTGAAACTCCTTCGACGAGTCGAGTTGTTTGGGAATGCAGCTCTAAATGGGTGGTAAATTTCATCTAAAGCTAAATATTGGCCAGAGACCGATAGCGCACAAGTAGAGTGATCGAAAGATGAAAAGCACTTTGAAAAGAGAGTTAAACAGTACGTGAAATTGTTGAAAGGGAAGCGCAGGCAACCAGACTTGCGCGTCGGGGTTCCCCCTTGCTTCTGCCTGGGTTACTCCCCGGCGTTCAGGCCAACATCGGTTTCGGGGGTTGGTTAAAGACCCTGGGAATGTATCTACCCTGCGGGGTAGACTTATAGCCCAGGGTGTCATGCGACCTCCCGGGACCGAGGAACGCGCTTCGGCTCGGATGTTGGCGTAATGGTTGTCTGCGGCCCGTCTTGAAACACGGACCAAGGAGTCTAACATCTATGCGAGTGTTAGGGTGTCAAACCCTTACGCGGAATGAAAGTGAACGGAGGTAGGAAGCCTTAAGGCTGCACTATCGACCGATCCTGAAGTTTACGGATGGATTTGAGTAAGAGCATAGCTGTTGGGACCCGAAAGATGGTGAACTATGCCTGAATAGGGTGAAGCCAGAGGAAACTCTGGTGGAGGCTCGCA >UAMH04935_JQ766402_RNA polymerase II AGCAGGTGAAAAAGATCCTAGAGACTGTCTGCCACAACTGTGGCAAAATCAAGGCACTTGAGGTTAGTAATACGTGATTCTAGTGATGCGCACTGCTCAGTTACTGACATTTCACAGGACGAGGCTTATCGACAAGCTCTTCGCGTTCGTAGTCGAAAGCTCCGATTCGAGCAGATTTGGCGCCTGTCCAAGAGCAGACTCGTCTGCGATGCCGACCCGCCTGAAGAAGGCGAGGGCGCCGCGAAACCTGGTCAAAATGTCATCAGACACGGCGGATGCGGCAACGTGCAGCCTGAGATGCGCAAGGCCGGCTTGCAGTTGTTCTCCACCTTTAAGCCCCGCAAGACAGACGATGACGACGACATGCCTGGAGGCAATCAGCCCGAGAAGAAGCGTTTCTTTGCTCAAGATGCTCTGAACGTCTTCCGCCTCATCTCCGATGAAACCCTTGAGACCCTCGGCTTGAGTGCCGACTATGCTCGACCAGAATGGATGATCCTCAACGCCCTTCCCGTACCACCGCCTCCAGTGCGTCCCAGTATCAGTGTCGACGGTAGTGGTCAGGGACAGCGTGGTGAAGATGATCTTACCTTCAAGCTCGGAGACATTATTCGCGCCAACCAGGCGGTCGAACGCACTGAATCGGATGGCACTCCTGACCACATCAAGATTGATTTGATCGACCTCCTGCA >UAMH04935_JQ766450_SSU-LSU CATTACCGAGTTAGGGTGTCAAACTTATGCGCCCGACCTCCAACCCTTTGTCTACTTGACCTTATGTTGCTTCGGCAGGCCCGCTGGTGTCCCTTTGGGGCCCGGCCGCCGGGGGTCCTCGTGGCCCCGGGTCCGCGCCTGCCGATGGCCCCTTCTTAAACTATTGTTAAAACGTGTCGTCTGAGTTTATGATTAAACAATCAAAACAAAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGCGAATTGCAGAATCCGTGAGTCATCGAATCTTTGAACGCACATTGCGCCCTTTGGTATTCCGAAGGGCATGCCTGTTCGAGCGTCATTATCACCCCTCAAGCCCGGCTTGTAGTTGGTTGCGGCGGCCCCTACCGCCCTTCCCAAAGATAATGACGGCGTCTGTGAGGACTCCTGTACACTGAGCTTTCGGGCACGTACACGGCTAGAAATCCAGGCCCGGTCGCCCTCGGCCCCCCTCGTTTTCGAGGGACCGAGAACCCCCTTTATATCAAGGTTGACC |
IMAGES:

