<< back to search
IMAGES:















Search Details
add to cart
| UAMH Number: | 10944 |
|---|---|
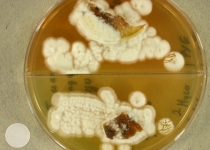
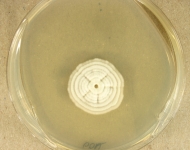
| Species Name: | Nannizziopsis guarroi |
| Type: | |
| Synonyms: | Chrysosporium guarroi |
| Taxonomy: | FUNGI Ascomycota, Eurotiomycetes, Onygenales, Onygenaceae |
| Strain History: | Gallego, M. (Tango 2) -> UAMH |
| Substrate: | leg green iguana (Iguana iguana) "Tango 2" | Location: | SPAIN Madrid, Centro Veterinario Madrid Exoticos (GEO: 40.433,-3.711) |
| Isolator: | M. Gallego |
| Isolation Date: | 2008-07-16 |
| Date Received: | 2008-09-09 |
| Characters: | HUMAN/ ANIMAL PATHOGEN deep mycosis in green iguana // MOLECULAR SYSTEMATICS phylogeny of Nannizziopsis and reptile pathogens belonging to the CANV complex - Sigler L, Hambleton S, Pare JA, J Clin Microbiol 51:3338-3357, 2013 // THERMOTOLERANT growth equivalent at 30 & 35C |
| Compounds: | |
| Cross Reference: | |
| Pathogenic Potential: | Human: no | Animal: yes | Plant: no |
| Biosafety Risk Group: | RG2 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) and a CFIA Written Authorization for transfer (http://www.inspection.gc.ca/plants/plant-pests-invasive-species/directives/date/d-12-03/eng/1432656209220/1432751554580#app2) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 801991 |
| Sequences: | >UAMH10944_KF477213_SSU-LSU CATTACTGTTGGTCACCGGGGGCCTCATCGCCCCCGGACCCTCCACCCGTGACTACTGAACATCCTTTGCCTCGGTCGGCCCAACGTGCTTGGAGCTCGCTTTCGGGCGCGCTCCGCCCACGGGGCCTGGATCCTGCGATCCGGGATGCGAAATCGCCGGCCGATGGCACCCCTGAACCTCTGTGAATAACGCAGTCTGAGTTTGATGTTAATCGTTAAAACTTTCAACAATGGATCTCTTGGTTCCCGCATCGATGAAGAACGCAGCGAAATGCGATAACTAATGTGAATTGCAGAATTCCGTGAATCATCGAGTCCTTGAACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTCCGAGCGTCATTGCACCCCTCAAGCGCAGCTTGGTGTTGGGCGACCGTCCCCCGGGACGCGCCCCAAATGCATTGGCGGCACCGGCACTGGAGCCCGGAGCGTATGGGAACCCCGTCACGCCCACAGGACCCGGCCGGAGCTGACCTTCAAATGATCGCTATTCATCCTCGGTTTGACCTCGGATCAGG |
IMAGES:













