Search Details
| UAMH Number: | 10475 |
|---|---|
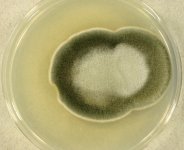
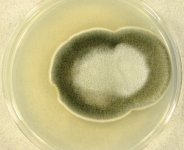
| Species Name: | Paradendryphiella salina |
| Type: | |
| Synonyms: | Cercospora salina / Dendryphiella salina / Scolecobasidium salinum |
| Taxonomy: | FUNGI Ascomycota, Dothideomycetes, Venturiales, Sympoventuriaceae |
| Strain History: | De la Cruz, T.E. (TUBs 7892) -> UAMH |
| Substrate: | red alga (Polysiphonia ureolata) | Location: | GERMANY Baltic Sea, Friedrichsort (GEO: 54.393,10.179) |
| Isolator: | T.E. De la Cruz (CPK 1862) |
| Isolation Date: | |
| Date Received: | 2004-09-13 |
| Characters: | BIODIVERSITY fungal secondary metabolites from marine habitats - Schulz B, Draeger S, de la Cruz TE, Rheinheimer J, Siems K, et al, Botanica Marina 51:219-234, 2008 // BIODIVERSITY marine species // CULTURE CONDITIONS growth responses to abiotic and biotic parameters found in marine habitats - De la Cruz TE, Wagner S, Schulz B, Mycol Progress 5:108-119, 2006 // CULTURE CONDITIONS utilization of carbon sources assessed with Biolog and API ZYM - De la Cruz TE, Schulz B, Kubicek CP, Druzhinina IS, FEMS Microbiol Ecol 58:343-353, 2006 |
| Compounds: | |
| Cross Reference: | BS05 |
| Pathogenic Potential: | Human: yes | Animal: yes | Plant: no |
| Biosafety Risk Group: | RG2 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) and a CFIA Written Authorization for transfer (http://www.inspection.gc.ca/plants/plant-pests-invasive-species/directives/date/d-12-03/eng/1432656209220/1432751554580#app2) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 803751 |
| Sequences: | >UAMH10475_DQ307336_elongation_factor-1_alpha ATTTTCTGAGCATGTAGCCATTTTCTGGCTTATCGCGATGAGGGGCATTTTTGGGTGGTGGGGTTGTGCGAACTTTTATGCGCTAGCGCTAGTCCGCATGTGGCCTTCGCGAACTGCAATACCATGACGCACATGCAAAATCCCATTTTCCGCTGCACGATGCTAACAGGCTCCATAGGAAGCCGCCGAGCTCGGTAAGGGTTCCTTCAAGTACGCATGGGTCCTCGACAAGTTGAAGGCTGAGCGTGAGCGTGGTATCACCATCGACATTGCTCTCTGGAAGTTCGAGACTCCCAAGTACTATGTCACCGTCATCGGTATGTTTCGCTCCCACCCTCCATTGCATCGCGCAGGTCTCTAACAGCAAGCAGACGCCCCCGGTCACCGTGATTTCATCAAGAACATGATCACTGGTACCTCCCAGGCCGACTGCGCTATTCTCATCATCGCTGCCGGTACTGGTGAGTTCGAGGCTGGTATCTCCAAGGACGGTCAGACTCGTGAGCACGCCCTGCTTGCCTACACCCTCGGTGTCAAGCAGCTCATCGTTGCCATCAACAAGATGGACACCACCAAGTGGTCTGAGGAGCGTTACCAGGAGATCATCAAGGAGACCTCCAACTTCATCAAGAAGGTCGGCTACAACCCCAAGCACGTTCCCTTCGTGCCCATCTCCGGTTTCAACGGTGACAACATGATTGAGGCCTCCACCAACTGCCCCTGGTACAAGGGTTGGGAGAAGGAGACCAAGTCCAAGGCTACCGGTAAGACCCTCCTCGAGGCCATCGACGCCATCGACCCTCCTAGCCGTCCTACCGACAAGCCCCTCCGCCTTCCCCTTCAAGATGTCTACAAGATTGGTGGGTATTGGCACGGTGCCTGTCGGTCGTGTCGAGAACCGGTGTCATCAAGGCCGGTATGGTCGTCACCTTCGCC >UAMH10475_DQ307292_ITS TGAAAGCGGGTTGGGACCTCATCTCGGTGGGGGCTCCAGCTTGTCTGAATTATTCACCCATGTCTTTTGCGCACTTCTTGTTTCCTGGGCGGGTTCGCCCGCCACCAGGACCCAACCATAAACCTTTTTTTGTAATTGCAATCAGCGTCAGTAAACAATGTAATTATTACAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATACGTAGTGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCTTTGGTATTCCAAAGGGCATGCCTGTTCGAGCGTCATTTGTACCCTCAAGCTTTGCTTGGTGTTGGGCGTCTTTGTCTCTCACGAGACTCGCCTTAAAATGATTGGCAGCCGGCCTACTGGTTTCGGAGCGCAGCACAATCTTGCACTTCTGATCAGCCATGGTTGAGCATCCATCAAGACCACATTTTTCTCACTTTTGACCTCGGATCAGGTAGGGATACCCGCTGAACTTAAGCATATCAATAAGCGGAG >UAMH10475_DQ307358_RNA_polymerase_II TACCTCCAGCGGTGTGTTGAGAACAACCAGGATTTCAACGTCCAGATGGCTGTCAAGGCTAGTATCATCACAAATGGATTGAAGTACTCTCTAGCGACAGGCAACTGGGGTGACCAGAAGAAGGCCGCGTCCGCCAAGGCCGGTGTCTCGCAGGTGTTGAACCGATACACTTACGCTTCCACACTTTCCCATCTTCGTCGAACAAATACTCCTGTTGGACGTGATGGTAAATTGGCTAAGCCGCGACAGCTCCACAACTCTCATTGGGGTCTTGTCTGCCCTGCCGAGACGCCTGAAGGACAGGCTTGCGGTCTGGTCAAGAACTTGTCCCTCATGTGCTATGTCAGTGTTGGTAGCGATGCATCGCCCATTATCGACTTCATGACTCAACGCAACATGCAACTTCTCGAGGAATACGACCAAAACCAAAATCCGGATGCGACCAAGGTCTTCGATCAACGGTGTTTGGGTTGGTGTTCATTCCAACGCTCAACAACTTGTCACAGTCGTACAGGAGCTTCGACGAAATGGCACCCTTTCCTACGAGATGAGTTTGATTCGTGATATTCGCGATCGAGAGTTCAAGATCTTCACAGATGCTGGCCGAGTCATGAGACCGCTGTTCGTCGTTGAGAAC |
IMAGES: