<< back to search
IMAGES:



Search Details
add to cart
| UAMH Number: | 10240 |
|---|---|
| Species Name: | Microascus cirrosus |
| Type: | |
| Synonyms: | |
| Taxonomy: | FUNGI Ascomycota, Sordariomycetes, Microascales, Microascaceae |
| Strain History: | Medlab -> Rogers, K. (MR 2341) -> UAMH |
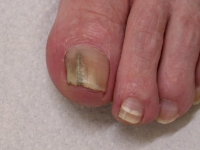
| Substrate: | nail male 60 yr; DE positive for nondermatophytic hyphae | Location: | NEW ZEALAND Hamilton (GEO: -37.787,175.279) |
| Isolator: | Medlab |
| Isolation Date: | 2016-02-10 |
| Date Received: | 2002-11-22 |
| Characters: | HUMAN/ ANIMAL PATHOGEN onychomycosis nails thickened with chalky black streaks, isolated twice // MOLECULAR SYSTEMATICS ITS sequence 1 bp different from UAMH 9389 T M. cirrosus // MOLECULAR SYSTEMATICS ITS sequence 100% identical to UAMH 10316 M. cirrosus // MOLECULAR SYSTEMATICS ITS sequence 100% identical to UAMH 9389 T M. cirrosus - fide UAMH 2011 |
| Compounds: | |
| Cross Reference: | |
| Pathogenic Potential: | Human: yes | Animal: no | Plant: no |
| Biosafety Risk Group: | RG1 (check the PHAC ePATHogen Risk Group Database for updates) |
| Regulatory Requirements: | Canadian requesters must provide PHAC Pathogen and Toxin License Number (see: https://www.canada.ca/en/public-health/services/laboratory-biosafety-biosecurity/licensing-program.html) prior to shipment. International requesters must provide all legally required importation documentation prior to shipment. Plant pathogenicity status may be verified by using the USDA Agricultural Research Service (ARS) Fungal Database |
| MycoBank ID: | 251450 |
| Sequences: | >UAMH10240_ITS CGATTGAATGGCTCAGTGAGGCCTTCGGACTGGCCCAGAGAGGCGGGCAACTGCCACTCAGGGCCGGAAAGTTGTCCAAACTCGGTCATTTAGAGGAAGTAAAAGTCGTAACAAGGTCTCCGTTGGTGAACCAGCGGAGGGATCATTACCGAAGTTACTCTTCATACCCTTTTGTGAACACTACCCCATTGCCGCGCGTTGCCTCGGCGGGGGAGGTCCGGGGGAGGGTTGCGCTCCGGCGCCTCCCCACCCTGGGCCCCCCGCCGGCCGCGCCGAACTCTTCATTTGCAAAGCGGACTGTACGTTCTGATTATATCTTGAAAAAACAAGTCAAAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCGGCAGCAATCTGCCGGGCATGCCTGTCCGAGCGTCATTTCTGCCCTCGAGCGCGGTTCGGTCCCTAGCGGGCCGGCCGCCGCCCGGTGTTGGGGCGCTGCGGGCCCTCGTGCCCGCAGGCCCTGAAATGAAGTGGCGGTCCCGCCGCGGCGCCCCCTGCGTAGTAGTAAAGCACCTCGCATCGGGACCCGGCGGAGGCCAGCCGTCGAACCTCTTCTATTGATGGT |
IMAGES:


